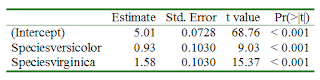
 Here's a quick tip for anyone wishing to export results, say a regression table, from R to MS Word:
Here's a quick tip for anyone wishing to export results, say a regression table, from R to MS Word:require(R2wd)
# install packages required
# install software RCOM, RDCOMClient
# (I had to restart the R-Session after the above step to get it work)
wdGet()
# Regression:
data(iris)
mod <- lm(Sepal.Length ~ Species, data = iris)
regr_tab <- data.frame(summary(mod)$coefficients)
colnames(regr_tab) <- colnames(summary(mod)$coefficients)
regr_tab[ ,4] <- ifelse(regr_tab[ ,4] < .001, "< 0.001",
ifelse(regr_tab[ ,4] < .01, "< 0.01",
round(regr_tab[ ,4], 3)))
# print table to doc in word-default format:
wdTable(format(regr_tab), autoformat = 1)
wdSave("Regression.doc") # save file
wdQuit() # close file
ps: Also check odfWeave
- this also seems to be a very useful resource!
Another way to do this is:
ReplyDeletemod <- lm(Sepal.Length ~ Species, data = iris)
library("xtable")
myXtable <- xtable( summary(mod) )
print(myXtable, type = "html")
The table will appear in HTML form. Cut and past that into any text editor and save it with an html extension. When you're done adding all your results to that file, save it, close it, then open the document using Word and choose whatever table formatting you like. To see all the objects that xtable supports, use:
methods(xtable)
thanks, for pointing out this method - for the export of tables only it seems less complicated to use xtable()!
Delete@Bob: I followed your suggestion but was unable to get the R output into a Word table. After opening the file in Word it simply reflects the copied output from R. What am I missing?
ReplyDeletenevermind -- got it to work. Thanks for sharing!
Delete..I had the same problem - how did you solve it?
Delete@ Bob and @ Key,
ReplyDeleteThanks for this post. Installing Rcom for the latest version of R was such a pain but it worked eventually. Building upon Bob's example, here is what i came up with. I have test this on R studio and it works flawlessly. thanks again.
regr_2_html <- function(reg_model, fname){
library("xtable")
library("R2HTML")
myXtable <- xtable( summary(reg_model) )
coef_rl = print(myXtable, type = "html")
# now here is the hack, i spit the file in html but with .doc ext so that it can be opened in word directly
HTML(coef_rl, paste(fname,"_model_coeff.doc", sep='')) ## this one works better
# write.table(coef_rl, paste(fname,"_model_coeff.html", sep='')) # can be used too
}
..thanks for sharing!
ReplyDeleteThanks a lot, you seem to be the only source that explains how to get a complete regression table.
ReplyDeleteDid anyone expand on this? Is there a how to which shows how to automatically include R² or something like that as well?
You're welcome! For R2 just add this line before saving to the doc-file:
Deleteregr_tab[,"R2"] <- c(summary(mod)$r.square, "", "")
Dear,
ReplyDeleteHere is a easy way to do that:
#install.packages('stargazer')
require(stargazer)
x <- rnorm(100,4,1)
y <- 3*x + rnorm(100,0,2)
model <- lm(y~x)
stargazer(model,type='html',
out='/user/reg.docx'
)
There is also a list of possible output styles. Check the help page of stargazer and you can find the options.
Best
Very cool - thanks!
Delete